Steve Sailer says he's "been alerted that there should be science news soon of a caliber comparable to the recent human-neanderthal inter-mating story." At Gene Expression, Greg Cochran calls attention to a comparison in the supplementary material of the Neanderthal genome paper showing "San closer to Han+French than to" Papuans. The San are also shown to be closer to the French than to the Chinese (and the Papuan closer to Han than French), though to a lesser degree. In addition, the San are shown to be closer to Han+French than to Yorubans, perhaps reflecting archaic admixture among West Africans; on the other hand, Yorubans are closer to non-Africans than to San.
Incidentally, this last point is in keeping with Kalinowski's assertion based on autosomal STR data. But as before I'm inclined to attribute any increased similarity between Negroids (as opposed to Bushmen) and non-Africans to back-migration from Eurasia to Africa, given that Y chromosomes likely of ultimate Eurasian origin (namely, E, and to a much lesser extent R1b, lineages) predominate among Negroids, while among Bushmen older clades predominate.
Update: A commenter passes on a link to a Google translation of a Spanish newspaper article reporting that Paabo and friends have sequenced the genome of the Denisova hominin, evidently a Homo erectus, and found evidence that genes related to the specimen persist in Melanesians. Update 2: Links are now dead (I assume the story is still supposed to be under embargo); see below for Google translation. Update 3: Much more information on this story is now available elsewhere, of course.
The computer program STRUCTURE does not reliably identify the main genetic clusters within species
Says this guy. While the author seems to be motivated by race-denialism, I think he's probably right that "forcing STRUCTURE to place individuals into too few clusters" can lead to non-optimal results (which would be consistent with what Dienekes appears to be finding). Certainly, contra Rienzi's railing against "incompetent amateurs, with their K=15 ADMIXTURE plots and clustering with up to 124 components", there's no reason to assume that in general runs with lower K will give more meaningful or accurate results than runs with higher K using programs like STRUCTURE and admixture.
I'm not terribly interested in Kalinowski's specific attempt to prove that African farmers are genetically closer to Europeans than to African hunter gatherers, but two things: (1) if this is true, I'd say it's much more likely to be attributable to back-migrations from Eurasia than to Europeans being descended from "African farmers"; (2) I enjoyed the section of the article in which the author promotes neighbor-joining trees as a superior method for visualizing relationships between populations, only to go on to explain that "even a tree with a R2 of 0.98 does not accurately depict all of the relationships between populations" since it still "depicts all sub-Saharan African populations as being more similar to each other than to European populations."
Reference: KALINOWSKI ST (2010) The computer program STRUCTURE does not reliably identify the main genetic clusters within species: simulations and implications for human population structure. Heredity (Published online). pdf
I'm not terribly interested in Kalinowski's specific attempt to prove that African farmers are genetically closer to Europeans than to African hunter gatherers, but two things: (1) if this is true, I'd say it's much more likely to be attributable to back-migrations from Eurasia than to Europeans being descended from "African farmers"; (2) I enjoyed the section of the article in which the author promotes neighbor-joining trees as a superior method for visualizing relationships between populations, only to go on to explain that "even a tree with a R2 of 0.98 does not accurately depict all of the relationships between populations" since it still "depicts all sub-Saharan African populations as being more similar to each other than to European populations."
Reference: KALINOWSKI ST (2010) The computer program STRUCTURE does not reliably identify the main genetic clusters within species: simulations and implications for human population structure. Heredity (Published online). pdf
Google ngrams
The Denisova hominin need not be an out of Africa story
I had a similar reaction when the original article was published, but this piece in the Journal of Human Evolution makes a much more extensive and better-argued case:
The recent retrieval of a complete mitochondrial (mt) DNA sequence from a 48–30 ka human bone from Denisova (Siberia) (Krause et al., 2010) is a remarkable achievement fully deserving international acclaim. Without wishing to detract from this feat, however, we wish to challenge their conclusion that the Denisova hominin “derives from a hominin migration out of Africa [ca. 1.0 Ma] distinct from that of the ancestors of Neanderthals and of modern humans” (Krause et al., 2010: 894). In addition, we challenge their assumption that the ancestors of the Neanderthals left Africa between 500–300 ka. In our view, alternative interpretations of the evidence are available and should be considered.Longer excerpts below:
China spying on deCODE?
Wikileaks: US Says Chinese Spies Operate in Iceland
Chinese authorities are believed to be spying on companies involved in genealogy and medical research in Iceland, as stated in documents sent by the US Embassy in Reykjavík to the US Foreign Service in Washington, leaked by Wikileaks.deCODE employs a Chinese national as "Vice President of Statistics", but Kari Steffanson is unconcerned on that front since the employee "has lived here for 14 years and considers Iceland his home".
This is stated in a report from February 26, 2009, which is marked as “confidential”. Copies of it were sent to the CIA, FBI and DIA, Fréttabladid reports.
In another report from December 24, 2009, also marked as “confidential”, an annual meeting of the US Embassy’s counter-spying group directed by the ambassador’s substitute Sam Watson is covered.
The report states that US authorities believe that their Chinese counterparts have continued with their intellectual spying in Iceland, by means of human intelligence and with technical equipment, such as bugging telephone lines and breaking into databases on the internet.
New England self-perception circa 1810
Some fairly random excerpts from Timothy Dwight's Travels in New England and New York (4 vols., 1821–1822).
Miscellaneous links
Polako: Locating and visualizing minority non-European admixtures across our genomes
Dienekes: Clusters galore: extremely fine-scale ancestry inference
Rienzi fumes about Dienekes' description of his own efforts as "cutting edge" and explains:
Hail: The Blackest Surnames in the USA; The Hated Richard Nixon’s Ancestry
Another theory on the origin of blond hair:
Dienekes: Clusters galore: extremely fine-scale ancestry inference
Rienzi fumes about Dienekes' description of his own efforts as "cutting edge" and explains:
In my opinion, Dienekes and Doug McDonald and Polako should be commended for attempting to further our understanding of human genetic diversity. They are trying to broaden our understanding. Hopefully, all three gentlemen are self-aware enough to know that their "findings" are at best tentative, should be interpreted cautiously, and await confirmation in the literature.This is not how science works. Anyone can download the software and reference data Dienekes is playing with and attempt to verify or impugn his results. Certainly some amateurs make ridiculous claims based on faulty analyses. So do some commercial entities and published academics. If you're not able to assess Dienekes' work on it's own merits, you're not in a better position to do so with comparable research merely because it has passed "peer review". One can certainly choose to ignore results generated by amateurs if one pleases, but choosing to do so has no bearing on the validity of the results.
Hail: The Blackest Surnames in the USA; The Hated Richard Nixon’s Ancestry
Another theory on the origin of blond hair:
Many traits have been investigated for their role in attractiveness, but one question has repeatedly captured the attention of researchers down the years – why do many men prefer blondes? Over time, many explanations have been put forward. It has been suggested that men prefer rounder faces and that blonde hair is kinder to the outline of the face, or that natural blondes have softer skin, which men find attractive. Another suggestion is that blondes were a genetic mutation which men evolved to value as a status symbol because of the original scarcity.Genetic Evidence for Multiple Biological Mechanisms Underlying In-Group Favoritism
But according to research out of the University of California, the answer is that blonde hair, like the peacock's tail or the rooster's bright-red plumage, is a sign of fitness. The evolutionary reason why men are attracted to blondes is that the hair and skin colour make it easier to spot problems. Anaemia, jaundice, skin infections, cyanosis (a sign of heart disease) and some other conditions, are, these researchers say, much easier to detect in fair-skinned individuals than in brunettes.
The best-fitting model revealed that a biological mechanism facilitates affiliation with arbitrary groups and exists alongside essentialist systems that evolved to process salient cues, such as shared beliefs and ancestry.
7 Ways the Mafia Made the U.S. a Better Place
TGGP drops a link to an article (republished by the ever-daring LewRockwell.com) by a HuffingtonPost contributor who fancies himself a purveyor of "Renegade History". Thaddeus Russell says America has Jewish and Italian gangsters to thank for:
- Jazz music
- Las Vegas
- Hollywood
- Racial mixing
- "Gay liberation"
23andMe sale today (24 November)
For those who are interested, according to someone on twitter:
DIYgenomics: @23andMe $99 discount returns; code B84YAG to be live 10 AM Wednesday for the new v3 chipThe updated chip is said to be the Illumina OmniExpress Plus:
The original press release described a chip with coverage of 733,202 markers, while the enhanced "Plus" version appears to cover greater than 900,000 markers. Either way, it is a significant upgrade from the previous 580,000 SNPs.Update: Discount code works. New chip tests 1,000,000+ SNPs.
Conferences
PDF slides of some presentations from Family Tree DNA's "6th International Conference on Genetic Genealogy", which took place at the end of October: Family Finder: Looking Under the Hood; Family Finder & Population Finder; "Inferring Genetic Ancestry: Oppourtunities, Challenges, and Implications"; IT Roadmap 2010; Predicting Individual Ancestry Using Genome-wide Genetic Data; Summarizing and Anticipating the Next Decade with NRY, mtDNA, and Autosomal DNA; Walk Through the Y Project.
Michael Hammer (according to an attendee): "village of origin can and will be done in the future as the database grows".
The 60th Annual ASHG meeting was held November 2-6 in Washington, D.C. A 23andMe employee comments and links to other coverage (see end of post) here. Video and slides from a 1000 Genomes Project tutorial here.
According to Luke Jostins:
Michael Hammer (according to an attendee): "village of origin can and will be done in the future as the database grows".
The 60th Annual ASHG meeting was held November 2-6 in Washington, D.C. A 23andMe employee comments and links to other coverage (see end of post) here. Video and slides from a 1000 Genomes Project tutorial here.
According to Luke Jostins:
at Biology of Genomes conference in May of next year; we’ll be putting out a large (~1100) sample dataset from around a dozen populations. These will be based on low-coverage whole-genome and high-coverage exome data on every sample, along with >2M genotypes from the Omni2.5 chip, to create a very high-quality set of data. Lots of work is going into putting together combined SNP, indel and CNV calls as nicely phased haplotypes. This dataset should be a massive boon to association studiesThis should also provide an additional public source of data for people undertaking projects like Polako's and Dienekes'.
CF mutant heterozygote advantage in heavy metal exposure
Teeth and leg bones from Iron Age people are showing a 21st century scientific-research team that there might be an evolutionary silver lining to the gene defects that cause cystic fibrosis (CF)
DNA analysis of ancient archeological finds is revealing that some CF gene defects may protect those who carry them from lead and other metal poisoning, or perhaps tuberculosis. [. . .]Via Jean M. The manuscript is freely available at Nature Precedings: Discovery of the Principal Cystic Fibrosis Mutation (F508del) in Ancient DNA from Iron Age Europeans
Since the protective CF gene mutation is so common among people living in or coming originally from central and Western Europe, Farrell suspects that the mutation first arose in that part of the world, very likely in early Celtic populations. [. . .]
To understand what in the environment could cause the mutated CF gene to occur in the first place, Farrell turned to ancient burial remains. Evidence from his earlier studies already showed that transgenic mice carrying the gene might be resistant to lead toxicity. He wanted to see if there were links to people living in Europe during the Iron and Bronze Ages.
“This was an era in which people were exposed to toxic heavy metals for the first time in history,” he says. [. . .]
The first analyses are showing that specimens containing CF gene defects were not affected by lead or other metal poisoning, hinting at the mutation’s protective advantage. The specimens also contained very little tuberculosis. The scientists can’t pinpoint exactly where the first CF carrier may have lived, but they think current day Austria is a good candidate.
Icelandic C1 distinct from Amerindian and Asian subclades
A new subclade of mtDNA haplogroup C1 found in icelanders: Evidence of pre-columbian contact?
Although most mtDNA lineages observed in contemporary Icelanders can be traced to neighboring populations in the British Isles and Scandinavia, one may have a more distant origin. This lineage belongs to haplogroup C1, one of a handful that was involved in the settlement of the Americas around 14,000 years ago. Contrary to an initial assumption that this lineage was a recent arrival, preliminary genealogical analyses revealed that the C1 lineage was present in the Icelandic mtDNA pool at least 300 years ago. This raised the intriguing possibility that the Icelandic C1 lineage could be traced to Viking voyages to the Americas that commenced in the 10th century. In an attempt to shed further light on the entry date of the C1 lineage into the Icelandic mtDNA pool and its geographical origin, we used the deCODE Genetics genealogical database to identify additional matrilineal ancestors that carry the C1 lineage and then sequenced the complete mtDNA genome of 11 contemporary C1 carriers from four different matrilines. Our results indicate a latest possible arrival date in Iceland of just prior to 1700 and a likely arrival date centuries earlier. Most surprisingly, we demonstrate that the Icelandic C1 lineage does not belong to any of the four known Native American (C1b, C1c, and C1d) or Asian (C1a) subclades of haplogroup C1. Rather, it is presently the only known member of a new subclade, C1e. While a Native American origin seems most likely for C1e, an Asian or European origin cannot be ruled out. Am J Phys Anthropol, 2010.The logic and evidence behind the authors' assertion that "a Native American origin seems most likely" is wanting; I find it much more likely C1 entered Iceland via Europe. Scientists will need to look elsewhere to explain Björk.
The 11 mutations that differentiate the Icelandic C1 sequences from the C1 root are in the upper range of mutation counts that differentiate the other C1 sequences from the root. [. . .]The frequency of C1 in a sample of 1538 Icelandic mtDNA sequences was 0.26%. Coincidentally, another abstract that recently appeared in PubMed is that of a Russian publication reporting:
A simple [polite way of saying retarded] argument in favor of a Native American origin of C1e is the fact that three of the four previously characterized C1 subclades are associated with these groups and the vast majority of C1 sequences in the literature have been sampled from individuals of Native American ancestry. [. . .]
The German sequence (Pfeiffer et al., 2001) represents a perfect match to the Icelandic C1e for the short HVS1 fragment spanning sites 16024–16365. This raises the intriguing, but perhaps unlikely, hypothesis that C1e is a European-specific subclade of C1, following the precedent of the European and Native American subclades of mtDNA haplogroup X2 (Brown et al., 1998; Reidla et al., 2003). However, given the dense sampling of mtDNA variation in European populations, it is clear that C1e is exceedingly rare, a fact that weighs against a hypothesis of antiquity in Europe.
The role of natural selection in the evolution of human populations from Northeastern Eurasia was studied. Selection for the regions-specific haplogroup C was demonstrated.
Update on People of the British Isles project
A reader forwarded me this message, posted to a mailing list by a third party:
One of my project members wrote to one of the organisers of the People of the British Isles Project to find out a few more details. He was told the following:The website is showing a total of 4214 samples collected.
"The data will be made publicly available after we have done some analyses, and so anyone should be able to get hold of it when it is!
As for SNPs, we have had 3,000 samples typed on a large scale (about 1.2M SNPs, of which something like 2,000 are on the Y-chromosome). There are about 150 or so that are on the y-chromosome consortium tree, so hopefully we should get quite a lot of information out of the analyses!"
That should make a big difference to Population Finder and all the other admixture tests. Perhaps those of us who already know that we are of 100% British origin might then actually get some meaningful results. The Orkney Islands are not exactly a good proxy for the entire British Isles!
Amerindian admixture in Gaspesia (Franch Canadia)
When Genetics and Genealogies Tell Different Stories-Maternal Lineages in Gaspesia
Data from uniparentally inherited genetic systems were used to trace evolution of human populations. Reconstruction of the past primarily relies on variation in present-day populations, limiting historical inference to lineages that are found among living subjects. Our analysis of four population groups in the Gaspé Peninsula, demonstrates how this may occasionally lead to erroneous interpretations. Mitochondrial DNA analysis of Gaspesians revealed an important admixture with Native Americans. The most likely scenario links this admixture to French-Canadians from the St. Lawrence Valley who moved to Gaspesia in the 19th century. However, in contrast to genetic data, analysis of genealogical record shows that Native American maternal lineages were brought to Gaspesia in the 18th century by Acadians who settled on the south-western coast of the peninsula. Intriguingly, within three generations, virtually all Métis Acadian families separated from their nonadmixed relatives and moved eastward mixing in with other Gaspesian groups, in which Native American maternal lines are present in relatively high frequencies. Over time, the carriers of these lines eventually lost memory of their mixed Amerindian-Acadian origin. Our results show that a reliable reconstruction of population history requires cross-verification of different data sources for consistency, thus favouring multidisciplinary approaches.I haven't read the article, so I have no idea on what basis the authors assert DNA results specifically pointed to "French-Canadians from the St. Lawrence Valley who moved to Gaspesia in the 19th century" as the most likely source of the admixture; but I'm all in favor of integrating DNA results with genealogical records in studies of this sort.
Ancestry analysis method
Jombart T, Devillard S, Balloux F. Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet. 2010 Oct 15;11(1):94.
BACKGROUND: The dramatic progress in sequencing technologies offers unprecedented prospects for deciphering the organization of natural populations in space and time. However, the size of the datasets generated also poses some daunting challenges. In particular, Bayesian clustering algorithms based on pre-defined population genetics models such as the STRUCTURE or BAPS software may not be able to cope with this unprecedented amount of data. Thus, there is a need for less computer-intensive approaches. Multivariate analyses seem particularly appealing as they are specifically devoted to extracting information from large datasets. Unfortunately, currently available multivariate methods still lack some essential features needed to study the genetic structure of natural populations.Website: http://adegenet.r-forge.r-project.org/
RESULTS: We introduce the Discriminant Analysis of Principal Components (DAPC), a multivariate method designed to identify and describe clusters of genetically related individuals. When group priors are lacking, DAPC uses sequential K-means and model selection to infer genetic clusters. Our approach allows extracting rich information from genetic data, providing assignment of individuals to groups, a visual assessment of between-population differentiation, and contribution of individual alleles to population structuring. We evaluate the performance of our method using simulated data, which were also analyzed using STRUCTURE as a benchmark. Additionally, we illustrate the method by analyzing microsatellite polymorphism in worldwide human populations and hemagglutinin gene sequence variation in seasonal influenza.
CONCLUSIONS: Analysis of simulated data revealed that our approach performs generally better than STRUCTURE at characterizing population subdivision. The tools implemented in DAPC for the identification of clusters and graphical representation of between-group structures allow to unravel complex population structures. Our approach is also faster than Bayesian clustering algorithms by several orders of magnitude, and may be applicable to a wider range of datasets.
Demographic simulation framework
Ray N, Currat M, Foll M, Excoffier L. SPLATCHE2: a spatially-explicit simulation framework for complex demography, genetic admixture and recombination. Bioinformatics. 2010 Oct 17.
SUMMARY: SPLATCHE2 is a program to simulate the demography of populations and the resulting molecular diversity for a wide range of evolutionary scenarios. The spatially-explicit simulation framework can account for environmental heterogeneity and fluctuations, and it can manage multiple population sources. A coalescent-based approach is used to generate genetic markers mostly used in population genetics studies (DNA sequences, SNPs, STRs, or RFLPs). Various combinations of independent, fully or partially linked genetic markers can be produced under a recombination model based on the ancestral recombination graph. Competition between two populations (or species) can also be simulated with user-defined levels of admixture between the two populations. SPLATCHE2 may be used to generate the expected genetic diversity under complex demographic scenarios and can thus serve to test null hypotheses. For model parameter estimation, SPLATCHE2 can easily be integrated into an Approximate Bayesian Computation (ABC) framework. Availability and Implementation: SPLATCHE2 is a C++ program compiled for Windows and Linux platforms. It is freely available at www.splatche.com, together with its related documentation and example data. CONTACT: mathias.currat@unige.ch.
Masculinity, skin color, and male facial attractiveness
Does Masculinity Matter? The Contribution of Masculine Face Shape to Male Attractiveness in Humans (PLoS ONE):
The proposal [. . .] that masculine men are immunocompetent and attractive – underpins a large literature on facial masculinity preferences. Recently, theoretical models have suggested that current condition may be a better index of mate value than past immunocompetence. This is particularly likely in populations where pathogenic fluctuation is fast relative to host life history. As life history is slow in humans, there is reason to expect that, among humans, condition-dependent traits might contribute more to attractiveness than relatively stable traits such as masculinity. [. . .]The authors point out problems with attempts to assess the affect of masculinity on facial attractiveness that rely on human ratings of perceived masculinity or digital manipulation of photographs: (1) for rated masculinity, "subjective judgments of masculinity are based on factors other than just morphological masculinity"; (2) with morphing techniques, factors potentially more important than masculinity in determining real world attractiveness are not allowed to vary, and a preference for averageness might result in participants systematically preferring more or less masculine morphs even if women are completely indifferent to masculinity. As for the effects of skin color, in this sample:
The relationship between masculinity and attractiveness was assessed in two samples of male faces. Most previous research has assessed masculinity either with subjective ratings or with simple anatomical measures. Here, we used geometric morphometric techniques to assess facial masculinity, generating a morphological masculinity measure based on a discriminant function that correctly classified >96% faces as male or female. When assessed using this measure, there was no relationship between morphological masculinity and rated attractiveness. In contrast, skin colour – a fluctuating, condition-dependent cue – was a significant predictor of attractiveness.
The regression retained only skin yellowness as a predictor of attractiveness, and the effect of skin yellowness was positive and highly significant (F(1,71) = 10.806, Beta = .366, t = 3.287, p<.002). Skin lightness, redness and morphological masculinity did not significantly predict attractiveness (all p>.114, see Table 1).Other studies have also found increased skin lightness and redness associated with perceived health and attractiveness. The association of yellowness with attractiveness "may be attributable to dietary carotenoid deposition in the skin. This suggests that carotenoids, which are involved in health signaling (Massaro et al. 2003; Saks et al. 2003) and sexual selection (Eley 1991; MacDougall and Montgomerie 2003; Massaro et al. 2003) in many species of birds and fish, may also affect the appearance of health in humans."
Race and physical attraction
A commenter links to a 2007 neuropolitics.org post ("Who Are The Caucasians Attracted To? Politics, Religion, and Physical Attraction") that reports the following survey results. As expected, "white females were more attracted to whites than are white males".
Miscellaneous links
Mangan on Transparency International's World Corruption Index
A One-Way Human Mission to Mars:
The origin of Eastern European Jews revealed by autosomal, sex chromosomal and mtDNA polymorphisms.
The Lost Tribes of Europe ("As national borders blur, the Continent's original minorities are fighting to reclaim their ancient cultures and identities")
Steve Sailer mentions: "a video of part of the amazing 1983 documentary First Contact with footage of the arrival of Australian explorers in the highlands of New Guinea around 1930."
A One-Way Human Mission to Mars:
There are many reasons why a human colony on Mars is a desirable goal, scientifically and politically. The strategy of one-way missions brings this goal within technological and financial feasibility. Nevertheless, to attain it would require [. . .] a return to the exploration spirit and risk-taking ethos of the great period of Earth exploration, from Columbus to Amundsen, but which has nowadays being replaced with a culture of safety and political correctness.
The origin of Eastern European Jews revealed by autosomal, sex chromosomal and mtDNA polymorphisms.
The Lost Tribes of Europe ("As national borders blur, the Continent's original minorities are fighting to reclaim their ancient cultures and identities")
Steve Sailer mentions: "a video of part of the amazing 1983 documentary First Contact with footage of the arrival of Australian explorers in the highlands of New Guinea around 1930."
Cranial differentiation of eastern and western pygmies
Diversity among African Pygmies (PLoS One):
Thirty three-dimensional (3D) landmarks registered with Microscribe in four cranial samples (Western and Eastern pygmies and non-pygmies) were obtained. Multivariate analysis (generalized Procrustes analysis, Mahalanobis distances, multivariate regression) and complementary dimensions of size were evaluated with ANOVA and post hoc LSD. Results suggest that important cranial shape differentiation does occur between pygmies and non-pygmies but also between Eastern and Western populations and that size changes and allometries do not affect similarly Eastern and Western pygmies. [. . .] Although not directly related to skull differentiation, the diversity among pygmies would probably suggest that the process responsible for reduced stature occurred after the split of the ancestors of modern Eastern and Western pygmies.
Middle-eastern milk drinkers?
A Der Spiegel article makes a case for the role of mass migration in shaping the European gene pool, while failing to mention the evidence for large-scale post-Neolithic population replacement. Jean M is unclear on whether the attempt to link lactase persistence to the LBK rests on unpublished aDNA results or questionable computer models. Greg Cochran on the latter eventuality:
Judging from the comment about lactose tolerance in Austria/Slovakia/Hungary, they may be relying on a paper that came out of Mark Thomas's lab last year: "The Origins of Lactase Persistence in Europe".John Hawks agrees:
The authors of that paper tried to estimate the region of origin using simulations - but one of the inputs was the current distribution of that allele. Which is reasonable, except that they did not use the actual distribution of that allele, but rather a truncated distribution - their map is centered on central Europe and stops halfway through the Ukraine. That ensured that they would find an origin in somewhere in middleuropa.
The allele doesn't stop there, though: it has a second region of fairly high frequency in northern India. Before Mongols and Turks took over the Eurasian steppe, the frequency of that allele may have been high in those steppe regions. Scythians are described as milk-drinkers quite a while ago - in the Iliad. And my sources claim that the royal guard of the Hittites also 'drank sweet milk'...
Checking out ancient DNA from Kurgan burials in that region might clarify this.
I think it is difficult to imagine a historical process that moves a lot of people from Bavaria to the Punjab: it is easier to imagine one that expands to both regions from somewhere in-between.
Which would explain the distribution of the Indo-European languages, also.
When you think about it, it may not be easy for German researchers to talk about this hypothesis. I think they have trouble saying "Aryan" nowadays.
Problem is: from the standpoint of ancient DNA samples, the lactase persistence mutation was also absent within the early Neolithic! The article is full of details that are wrong or misleading. [. . .]
The [mtDNA] differences between early Neolithic and later Europeans suggests that post-Neolithic migrations -- real Völkerwandurung -- actually had a major impact on the European gene pool. What we see today is not a pattern established 6000 years ago, but a palimpsest richly painted with strokes from successive migrations.
One aspect of this scenario: There's no reason to link the early Neolithic with Indo-European languages. There were many later widespread population movements that might have carried this language family, and we know that these later movements were genetically decisive -- at least, as concerns the maternal genealogy. The relation of Y chromosome haplogroups with mtDNA haplogroups is a critical question, but even more necessary is the development of an effective means of testing these hypotheses with nuclear genotype data.
Miscellaneous links
Bio-IT World road to $1,000 genome issue (via Genomes Unzipped)
Distinct clones of Yersinia pestis Caused the Black Death (via Jean M)
Inference of Unexpected Genetic Relatedness among Individuals in HapMap Phase III
"non-western children don’t do so well on the old mirror-recognition test"
"Lost" Francis Crick correspondence found
Darwin's Family Tree Re-Discovered
Distinct clones of Yersinia pestis Caused the Black Death (via Jean M)
Inference of Unexpected Genetic Relatedness among Individuals in HapMap Phase III
"non-western children don’t do so well on the old mirror-recognition test"
"Lost" Francis Crick correspondence found
Wilkins [. . .] complained to Crick and Watson in a rediscovered letter: "To think that Rosie had all the 3D data for 9 months & wouldn’t fit a helix to it and there was I taking her word for it that the data was anti-helical. Christ."A rare variant of the mtDNA HVS1 sequence in the hairs of Napoleon's family
Darwin's Family Tree Re-Discovered
The horse, the wheel, and language
It appears someone took the liberty of uploading David Anthony's book on Indo-European origins to scribd. Read and discuss if you wish.
Genetic deterioration of modern populations?
A post by Notus Wind brought this paper to my attention:
Although mutation provides the fuel for phenotypic evolution, it also imposes a substantial burden on fitness through the production of predominantly deleterious alleles, a matter of concern from a human-health perspective. [. . .] a consideration of the long-term consequences of current human behavior for deleterious-mutation accumulation leads to the conclusion that a substantial reduction in human fitness can be expected over the next few centuries in industrialized societies unless novel means of genetic intervention are developed.I know Hamilton expressed similar concerns, but to the extent accelerated accumulation of deleterious mutations under modern conditions is a real/serious problem this author's suggestion of "multigenerational cryogenic storage and utilization of gametes and/or embryos" seems preferable to some of Hamilton's goofier "solutions" (such as marrying HIV- Nairobi prostitutes to protect one's offspring from impending AIDS epidemic). I'm not too worried about imminent mutational meltdown (some modern "problems" like antibiotics may be in the process of solving themselves), but from a strictly conservationist or even historical standpoint, large-scale, long-term storage of human genetic and/or gametic material makes sense. If we can see the need for plants and animals, why not for ourselves?
Ptolemy map of ancient Germania deciphered
Berlin Researchers Crack the Ptolemy Code
All this offers up rather exciting prospects, since it makes half the cities in Germany suddenly 1,000 years older than previously believed. "Our atlas is a treasure map," team member Andreas Kleineberg says proudly, "and the coordinates lead to lost places in our past."
Archaeological interest in the map will likely be correspondingly large. Archaeologists' opinions on the Germanic tribes have varied over the years. In the 19th century, Germany's early inhabitants were considered brave, wild-bearded savages. The Nazis then transformed them into great heroes, and in the process of coming to terms with its Nazi past, postwar Germany quickly demoted the early Germanic peoples to proto-fascist hicks. [. . .] More recent research proves this view to be complete invention.
Charlemagne was very tall, but not robust
Paper mentioned by John Hawks: "Reconstructed stature of 1.84 m falls at about 99% of Medieval heights [. . .] Thus, tall stature indeed could have contributed to the success of “Charles the Great” as a king emperor and soldier."
Below, a couple other papers that appeared in Hawks's citeulike feed.
The biological standard of living in Europe during the last two millennia: "We find that heights stagnated in Central, Western and Southern Europe during the Roman imperial period, while astonishingly increasing in the fifth and sixth centuries."
New Light on the "Dark Ages": The Remarkably Tall Stature of Northern European Men during the Medieval Era: "Based on a modest sample of skeletons from northern Europe, average heights fell from 173.4 centimeters in the early Middle Ages to a low of roughly 167 centimeters during the seventeenth and eighteenth centuries. [. . .] It is plausible to link the decline in average height to climate deterioration; growing inequality; urbanization and the expansion of trade and commerce, which facilitated the spread of diseases; fluctuations in population size that impinged on nutritional status; the global spread of diseases associated with European expansion and colonization; and conflicts or wars over state building or religion."
Below, a couple other papers that appeared in Hawks's citeulike feed.
The biological standard of living in Europe during the last two millennia: "We find that heights stagnated in Central, Western and Southern Europe during the Roman imperial period, while astonishingly increasing in the fifth and sixth centuries."
New Light on the "Dark Ages": The Remarkably Tall Stature of Northern European Men during the Medieval Era: "Based on a modest sample of skeletons from northern Europe, average heights fell from 173.4 centimeters in the early Middle Ages to a low of roughly 167 centimeters during the seventeenth and eighteenth centuries. [. . .] It is plausible to link the decline in average height to climate deterioration; growing inequality; urbanization and the expansion of trade and commerce, which facilitated the spread of diseases; fluctuations in population size that impinged on nutritional status; the global spread of diseases associated with European expansion and colonization; and conflicts or wars over state building or religion."
Preferred and actual mate characteristics
From Preferred to Actual Mate Characteristics: The Case of Human Body Shape
The way individuals pair to produce reproductive units is a major factor determining evolution. This process is complex because it is determined not only by individual mating preferences, but also by numerous other factors such as competition between mates. Consequently, preferred and actual characteristics of mates obtained should differ, but this has rarely been addressed. We simultaneously measured mating preferences for stature, body mass, and body mass index, and recorded corresponding actual partner's characteristics for 116 human couples from France. Results show that preferred and actual partner's characteristics differ for male judges, but not for females. In addition, while the correlation between all preferred and actual partner's characteristics appeared to be weak for female judges, it was strong for males: while men prefer women slimmer than their actual partner, those who prefer the slimmest women also have partners who are slimmer than average. This study therefore suggests that the influences of preferences on pair formation can be sex-specific. It also illustrates that this process can lead to unexpected results on the real influences of mating preferences: traits considered as highly influencing attractiveness do not necessarily have a strong influence on the actual pairing, the reverse being also possible.
2010 Forbes 400 by ancestry: more statistics
See preceding post for complete list.
| Net Worth | No. of child- ren | ||||||
| n | Total billions | Pct of total | Mean | Median | Age | ||
| NW Euro | 203 | 764.45 | 55.84 | 3.77 | 2 | 67.24 | 3.19 |
| Jewish | 143 | 486.5 | 35.54 | 3.4 | 1.8 | 63.44 | 3.06 |
| Italian | 17 | 34.9 | 2.55 | 2.05 | 1.6 | 61.76 | 2.71 |
| E Asian | 8 | 22.4 | 1.64 | 2.8 | 2.6 | 60.63 | 2.25 |
| Mid East | 8 | 22.1 | 1.61 | 2.76 | 1.95 | 72 | 3.13 |
| E Euro | 7 | 16.2 | 1.18 | 2.31 | 2.5 | 68.86 | 3.43 |
| Greek | 7 | 11.5 | 0.84 | 1.64 | 1.7 | 77.57 | 4 |
| S Asian | 4 | 5.75 | 0.42 | 1.44 | 1.43 | 57 | 2.25 |
| Hispanic | 2 | 2.4 | 0.18 | 1.2 | 1.2 | 68.5 | 2.5 |
| Black | 1 | 2.7 | 0.2 | 2.7 | 2.7 | 56 | 0 |
| Overall | 400 | 1368.9 | 100 | 3.42 | 2 | 65.7 | 3.1 |
2010 Forbes 400 by ethnic origins
[Related: 2012 Forbes 400 by ethnic origins; more statistics on 2010 Forbes 400 by ancestry.]
The 2009 estimate is also mine, and I have used the same methodology as before. The 1987 estimate is Nathaniel Weyl's. The first thing I notice is that the Northwestern European proportion of the list continues to decline.
Full list below. Feel free to post corrections.
[Correction: Moved Frost to Jewish section; updated statistics accordingly.]
Forbes has published its list of the 400 richest Americans for this year. My current estimate of the ethnic breakdown of the list is shown in the far right column below:
| 1987 (%) | 2009 (%) | 2010 (%) | |
| Northwestern European | 72 | 52.25 | 51 |
| Jewish | 23 | 35 | 35 |
| Italian | 2.25 | 3.5 | 4.25 |
| East Asian | 0.25 | 2.0 | 2.0 |
| Middle Eastern | 1.5 | 2.25 | 2.5 |
| Greek | 0.5 | 1.5 | 1.75 |
| Eastern European | 0.25 | 1.5 | 1.75 |
| South Asian | 0.0 | 1.25 | 1.0 |
| Hispanic | 0.25 | 0.5 | 0.5 |
| Black | 0.0 | 0.25 | 0.25 |
Full list below. Feel free to post corrections.
"Thousands" of genes influence intelligence
A commenter writes: "Robert Plomin and colleagues have apparently found some IQ genes". From the story:
SCIENTISTS have identified more than 200 genes potentially associated with academic performance in schoolchildren. [. . .]So much for the fantasies of certain technorapturist HBDers that genetic engineering will erase racial differences in intelligence within a few decades.
The finding emerged from a study of more than 4000 British children to pinpoint the genes and genetic combinations that influence reasoning skills and general intelligence.
One of its main conclusions is that intelligence is controlled by a network of thousands of genes with each making just a small contribution to overall intelligence, rather than the handful of powerful genes that scientists once predicted.
Population structure math: two recent papers
Analysis of Population Structure: A Unifying Framework and Novel Methods Based on Sparse Factor Analysis
Two different approaches have become widely used in the analysis of population structure: admixture-based models and principal components analysis (PCA). In admixture-based models each individual is assumed to have inherited some proportion of its ancestry from one of several distinct populations. PCA projects the individuals into a low-dimensional subspace. On the face of it, these methods seem to have little in common. Here we show how in fact both of these methods can be viewed within a single unifying framework. This viewpoint should help practitioners to better interpret and contrast the results from these methods in real data applications. It also provides a springboard to the development of novel approaches to this problem. We introduce one such novel approach, based on sparse factor analysis, which has elements in common with both admixture-based models and PCA. As we illustrate here, in some settings sparse factor analysis may provide more interpretable results than either admixture-based models or PCA.Theoretical Formulation of Principal Components Analysis to Detect and Correct for Population Stratification
The Eigenstrat method, based on principal components analysis (PCA), is commonly used both to quantify population relationships in population genetics and to correct for population stratification in genome-wide association studies. However, it can be difficult to make appropriate inference about population relationships from the principal component (PC) scatter plot. Here, to better understand the working mechanism of the Eigenstrat method, we consider its theoretical or “population” formulation. The eigen-equation for samples from an arbitrary number () of populations is reduced to that of a matrix of dimension , the elements of which are determined by the variance-covariance matrix for the random vector of the allele frequencies. Solving the reduced eigen-equation is numerically trivial and yields eigenvectors that are the axes of variation required for differentiating the populations. Using the reduced eigen-equation, we investigate the within-population fluctuations around the axes of variation on the PC scatter plot for simulated datasets. Specifically, we show that there exists an asymptotically stable pattern of the PC plot for large sample size. Our results provide theoretical guidance for interpreting the pattern of PC plot in terms of population relationships. For applications in genetic association tests, we demonstrate that, as a method of correcting for population stratification, regressing out the theoretical PCs corresponding to the axes of variation is equivalent to simply removing the population mean of allele counts and works as well as or better than the Eigenstrat method.
Assortative mating by ancestry among American whites
Testing for non-random mating: evidence for ancestry-related assortative mating in the Framingham heart study.
Population stratification leads to a predictable phenomenon-a reduction in the number of heterozygotes compared to that calculated assuming Hardy-Weinberg Equilibrium (HWE). We show that population stratification results in another phenomenon-an excess in the proportion of spouse-pairs with the same genotypes at all ancestrally informative markers, resulting in ancestrally related positive assortative mating. We use principal components analysis to show that there is evidence of population stratification within the Framingham Heart Study, and show that the first principal component correlates with a North-South European cline. We then show that the first principal component is highly correlated between spouses (r = 0.58, p = 0.0013), demonstrating that there is ancestrally related positive assortative mating among the Framingham Caucasian population. We also show that the single nucleotide polymorphisms loading most heavily on the first principal component show an excess of homozygotes within the spouses, consistent with similar ancestry-related assortative mating in the previous generation. This nonrandom mating likely affects genetic structure seen more generally in the North American population of European descent today, and decreases the rate of decay of linkage disequilibrium for ancestrally informative markers. Genet. Epidemiol. 2010.
Sub-Saharan mtDNA and malaria in Portugal
Genetic characterization of uniparental lineages in populations from Southwest Iberia with past malaria endemicity:
Malaria endemicity in Southwest Iberia afforded conditions for an increase of sickle cell disease (SCD), which in the region follows a clinal pattern toward the south, where foci of high prevalence were found. SCD distribution is associated with specific geographical areas, and therefore, its introduction into Iberia may be related to the migration of different populations. We have analyzed the variation of uniparental markers in Portuguese populations with high frequency of SCD-Coruche, Pias, and Alcacer do Sal-to evaluate if their present-day pattern of neutral diversity could provide evidence about people inhabiting the area over different time periods. Two hundred and eighty-five individuals were sampled in Coruche, Pias, and Alcacer do Sal. All were analyzed for the control region of mitochondrial DNA (mtDNA); males were additionally examined for Y-chromosome markers. Results were then compared with data from other Portuguese and non-Portuguese populations. In Coruche, the genetic profile was similar to the profile usually found in Portugal. In Alcacer do Sal, the frequency of sub-Saharan mtDNA L lineages was the highest ever reported (22%) in Europe. In Pias, mtDNA diversity revealed higher frequencies of Mediterranean haplogroups I, J, and T than usually found in surrounding populations. The presence of Sub-Saharan maternal lineages in Alcacer do Sal is likely associated with the influx of African slaves between the 15th and 19th centuries, whereas in Pias, the Mediterranean influence might be traced to ancient contacts with Greeks, Phoenicians, and Carthaginians, who established important trading networks in southern Iberia. Am. J. Hum. Biol. 22:588-595, 2010.Women selling seafood in Alcácer do Sal.
"WASP" decline: NYG&B edition
3 Years After a Windfall, an Unexpected Reality for the Genealogical Society:
Since 1869, the New York Genealogical and Biographical Society has made its name helping New Yorkers understand their roots. But the more the society pondered its future, the more the people at its helm became convinced that it needed to break with its past.The new owners: a synagogue. In a comment, the new NYGBS president insists:
Making what they called a “forward-looking decision,” the trustees hung a for-sale sign on the society’s longtime home at 122 East 58th Street several years ago. Then — to the shock of the society’s own members, who were still reeling from the sale — the trustees gave the society’s vast collection of scholarly books and historic papers to the New York Public Library.
The radical downsizing may have been as pragmatic as it was bold, but the timing could have been better.
The building was sold in 2007, and one-fourth of the $24 million that the society raised from the sale washed away as the stock market eroded. That, together with high administrative costs, led the society to make large staff reductions in early 2009, just as the new owners of its old headquarters sold them again — this time for $28.5 million.
there is no doubt whatever that the choices were carefully, thoroughly, and thoughtfully considered by dedicated trustees and by the consultants who advised them over an extended period of time.I don't doubt most of the trustees were doing what they thought was best. But I'd be curious to know the names of the consultants involved.
Miscellaneous links
Unpublished 1947 photos of the Lascaux cave paintings (via hbdchick)
German woman gets to thank Portland woman in person for post-WWII CARE packages
Genetic contribution to variation in body configuration in Belgian nuclear families: a closer look at body lengths and circumferences
1000 Genomes Project: Y Chromosome SNPs (pdf)
Extended Y-chromosome investigation suggests post-Glacial migrations of modern humans into East Asia via the northern route ("evidence of geographic distribution and Y-STR diversity indicate that haplogroup Q-M242 (the ancestral haplogroup of the native American-specific haplogroup Q1a3a-M3) and R-M207 probably migrated into East Asia via the northern route")
Genetic Differences between Five European Populations ("several genes have not previously been reported as stratified within European populations, indicating that they might also have provided selective advantages: [including] most intriguingly, FOXP2, implicated in speech development.")
German woman gets to thank Portland woman in person for post-WWII CARE packages
Genetic contribution to variation in body configuration in Belgian nuclear families: a closer look at body lengths and circumferences
1000 Genomes Project: Y Chromosome SNPs (pdf)
Extended Y-chromosome investigation suggests post-Glacial migrations of modern humans into East Asia via the northern route ("evidence of geographic distribution and Y-STR diversity indicate that haplogroup Q-M242 (the ancestral haplogroup of the native American-specific haplogroup Q1a3a-M3) and R-M207 probably migrated into East Asia via the northern route")
Genetic Differences between Five European Populations ("several genes have not previously been reported as stratified within European populations, indicating that they might also have provided selective advantages: [including] most intriguingly, FOXP2, implicated in speech development.")
Additional ASHG 2010 abstracts
Selected abstracts from the 2010 meeting of the American Society of Human Genetics.
ASHG 2010: Ancestry and admixture
Selected abstracts from the 2010 meeting of the American Society of Human Genetics.
ASHG 2010: Personal genomics
Selected abstracts from the 2010 meeting of the American Society of Human Genetics.
ASHG 2010: Pigmentation genetics
Selected abstracts from the 2010 meeting of the American Society of Human Genetics.
ASHG 2010: Adaptation and natural selection
Selected abstracts from the 2010 meeting of the American Society of Human Genetics.
ASHG 2010: South Asian genetics
Selected abstracts from the 2010 meeting of the American Society of Human Genetics.
Autosomal African admixture in Yemeni populations
Another ASHG 2010 abstract. Using CEU and Maasai reference populations, the authors estimate Yemenis average about 14.8% African admixture.
ASHG 2010 abstracts on Jewish genetics
Selected abstracts from the 2010 meeting of the American Society of Human Genetics.
ASHG 2010 abstract on archaic admixture in modern humans
Search abstracts for the upcoming meeting of the American Society of Human Genetics here.
Evidence for archaic admixture in contemporary non-African human populations. J. Long, R. Ferrucci, S. Joyce, K. Hunley Dept of Anthropology, Univeristy of New Mexico, Albuquerque, NM.
New, open access genetics journal
Investigative Genetics. Some of the areas of interest mentioned: forensic genetics, identity and lineage testing, personalized genetics, population genetics, evolutionary genetics, mechanisms of inheritance, ancient biomaterials, anthropological genetics, archaeogenetics. Mark Jobling will have a regular column. The inaugural issue includes the following paper.
Development of a single base extension method to resolve Y chromosome haplogroups in sub-Saharan African populations
Development of a single base extension method to resolve Y chromosome haplogroups in sub-Saharan African populations
the assays were used to screen 683 individuals from Southern Africa, including south eastern Bantu speakers (BAN), Khoe-San (KS) and South African Whites (SAW). Of the 61 haplogroups that the assays collectively resolved, 26 were found in the 683 samples. While haplogroup sharing was common between the BAN and KS, the frequencies of these haplogroups varied appreciably. Both groups showed low levels of assimilation of Eurasian haplogroups and only two individuals in the SAW clearly had Y chromosomes of African ancestry.
Redhead day 2010
Wikipedia: "Redheadday is the name of a Dutch summer festival that takes place each first weekend of September in the city of Breda, in the Netherlands."
Facts about redheads: "Red hair is seen on the heads of only less than one percent of people.in the world. Most redheads live in the U.K., Ireland, and former colonies of U.K. like Australia.
The highest percentage of natural Redheads in the world is in Scotland (13%), followed closely by Ireland with 10%. In the US, about 2% of the population are natural redheads.
Redheads are becoming rarer and could be extinct in 100 years, according to genetic scientists."
(Photo credit. More photographs.)
Facts about redheads: "Red hair is seen on the heads of only less than one percent of people.in the world. Most redheads live in the U.K., Ireland, and former colonies of U.K. like Australia.
The highest percentage of natural Redheads in the world is in Scotland (13%), followed closely by Ireland with 10%. In the US, about 2% of the population are natural redheads.
Redheads are becoming rarer and could be extinct in 100 years, according to genetic scientists."
(Photo credit. More photographs.)
Free Access to Sage Journals until Oct 15th
More information here: "The next generation of SAGE Journals Online (SJO), SAGE's award-winning journal platform, is now live! To celebrate, free online access to must-have content back to 1999 is available until October 15, 2010" (via Jason Malloy).
David Sloan Wilson on kin selection and group selection
Open Letter to Richard Dawkins: Why Are You Still In Denial About Group Selection?
I do not agree with the Nowak et al. article in every respect and will articulate some of my disagreements in subsequent posts. For the moment, I want to stress how alone you are in your statement about group selection. Your view is essentially pre-1975, a date that is notable not only for the publication of Sociobiology but also a paper by W.D. Hamilton, one of your heroes, who correctly saw the relationship between kin selection and group selection thanks to the work of George Price. Ever since, knowledgeable theoretical biologists have known that inclusive fitness theory includes the logic of multilevel selection, which means that altruism is selectively disadvantageous within kin groups and evolves only by virtue of groups with more altruists contributing more to the gene pool than groups with fewer altruists. The significance of relatedness is that it clusters the genes coding for altruistic and selfish behaviors into different groups.See earlier post for links to replies to Nowak et al. by Dawkins and others.
New(ish) blogs
Ahnenkult. According to the about page:
The Anthroconservative Beacon. "A Daily Blog Dedicated to Upholding Conservative Principles Based on a Scientific Understanding of Human Nature" by "an Associate Professor in the State University of New York system."
HBD Talk. "Political Views from a Secular Humanist Perspective". Some have suggested this is Richard Hoste's new blog. I couldn't say. I skipped all the posts except the one on African blood in Arabs [Update: The person behind HBD Talk says he is not Hoste].
This blog is concerned with history, both human and natural, and race in its various senses. [] Ortu Kan is an Asiatic in the second decade of his life, temporarily in the West but not of it. His people’s language is Altaic (sensu Miller 1971).Posts include: 'Were there Negritos on Taiwan?', 'Ushtettas as Tunisian "Australoids"', 'Cave-Dwelling Click Speakers in 16th-Century Mozambique'
The Anthroconservative Beacon. "A Daily Blog Dedicated to Upholding Conservative Principles Based on a Scientific Understanding of Human Nature" by "an Associate Professor in the State University of New York system."
HBD Talk. "Political Views from a Secular Humanist Perspective". Some have suggested this is Richard Hoste's new blog. I couldn't say. I skipped all the posts except the one on African blood in Arabs [Update: The person behind HBD Talk says he is not Hoste].
Miscellaneous links
HapMap 3 Consortium: Integrating common and rare genetic variation in diverse human populations
African signatures of recent positive selection in human FOXI1:
Distinct patterns of mitochondrial genome diversity in bonobos (Pan paniscus) and humans:
Distinct genomic alterations in prostate cancers in Chinese and Western populations suggest alternative pathways of prostate carcinogenesis
How many journal articles have been published (ever)? / Twenty million papers in PubMed: a triumph or a tragedy?
Ancestral Health Symposium (August 5-6, 2011)
African signatures of recent positive selection in human FOXI1:
Conclusions: We present evidence for recent positive selection in the FOXI1 gene region in Africa. Climate might be related to this recent adaptive event in humans. Of the multiple functions of FOXI1, its role in kidney-mediated water-electrolyte homeostasis is the most obvious candidate for explaining a climate-related adaptation.Y Chromosomal Variation Tracks the Evolution of Mating Systems in Chimpanzee and Bonobo
Distinct patterns of mitochondrial genome diversity in bonobos (Pan paniscus) and humans:
Conclusions: Some variants of mitochondrially encoded subunits of the ATPase complex in humans very likely decrease the efficiency of energy conversion leading to production of extra heat.Evolution Theory as the “Consilience” Ingredient of the Humanities:
The Humanities are awash with conflicting, baseless explanations of Human Nature. Sociology, Psychology, Rational Choice theory, Religion, Marxism, Feminisim–all fail in their EXPLANATORY power when tested for consistency when they butt up against the observable actions of real human beings on the Planet. [. . .] In his article Take The Evolution Challenge, David Sloan Wilson eloquently proposes that Evolution Theory be used as a Universal Solvent for all of these competing theories, burning away the accretions of years of constructivist and ideological fat and uncovering the truth.How do we choose a mate? What scientists are learning from online dating.
Distinct genomic alterations in prostate cancers in Chinese and Western populations suggest alternative pathways of prostate carcinogenesis
How many journal articles have been published (ever)? / Twenty million papers in PubMed: a triumph or a tragedy?
Ancestral Health Symposium (August 5-6, 2011)
Kin selection not dead
Explaining eusociality:
There's a slightly embarrassing paper out in Nature last week with E.O. Wilson on the list of authors that disparages inclusive fitness (the logic behind kin selection) and insists that (a) its explanatory power and contributions to evolutionary biology have been limited and (b) it can be ditched outright in favor of "standard natural selection theory". What is meant by this last phrase is not entirely clear, but what is clear is that the authors do not understand inclusive fitness and have completely overlooked the many ways in which it has illuminated us (sex ratio adjustment as the prime example). The authors have a section titled "Rise and fall of inclusive fitness theory", which is curious unless they are writing alt history fiction.John Hawks agrees:
I am only going to comment briefly on the paper, because Jerry Coyne and Richard Dawkins have already chewed it up.
The weird part of the paper is the way it describes inclusive fitness as some kind of theoretical afterthought, useful only as an ad hoc explanation for eusocial insects. It contrasts the inclusive fitness concept with "standard natural selection" as if it were possible for organisms to erase the fact that they're related to each other! And the authors imply that they have fatally damaged the concept of kin selection.
It's so contrary to evolutionary theory, that I thought maybe I was missing something. But I've been spending time on another problem this week and haven't had time to follow it up.
Two new papers on the distribution of R1b subclades
Myres et al. (pdf):
The dating (and consequently the authors' specific attempts to link the spread of R1b to archaeological horizons) is likely badly off; the various subclades are probably much younger than calculated by the authors. It's also far from clear to me that R1b entered Europe via Anatolia (as opposed to a more northerly route). Either way, the Iberian Irish myth is still very dead.
I haven't read the Cruciani paper (pdf).
References:
Myres et al. A major Y-chromosome haplogroup R1b Holocene era founder effect in Central and Western Europe. European Journal of Human Genetics doi: 10.1038/ejhg.2010.146
Cruciani et al. Strong intra- and inter-continental differentiation revealed by Y chromosome SNPs M269, U106 and U152. Forensic Science International: Genetics doi:10.1016/j.fsigen.2010.07.006
present phylogeographically resolved data for 2043 M269-derived Y-chromosomes from 118 West Asian and European populations assessed for the M412 SNP that largely separates the majority of Central and West European R1b lineages from those observed in Eastern Europe, the Circum-Uralic region, the Near East, the Caucasus and Pakistan. Within the M412 dichotomy, the major S116 sub-clade shows a frequency peak in the upper Danube basin and Paris area with declining frequency toward Italy, Iberia, Southern France and British Isles.The supplementary data contains more detailed information on the frequency and diversity of R1b subclades in various population samples. Dienekes comments.
The dating (and consequently the authors' specific attempts to link the spread of R1b to archaeological horizons) is likely badly off; the various subclades are probably much younger than calculated by the authors. It's also far from clear to me that R1b entered Europe via Anatolia (as opposed to a more northerly route). Either way, the Iberian Irish myth is still very dead.
I haven't read the Cruciani paper (pdf).
References:
Myres et al. A major Y-chromosome haplogroup R1b Holocene era founder effect in Central and Western Europe. European Journal of Human Genetics doi: 10.1038/ejhg.2010.146
Cruciani et al. Strong intra- and inter-continental differentiation revealed by Y chromosome SNPs M269, U106 and U152. Forensic Science International: Genetics doi:10.1016/j.fsigen.2010.07.006
Signatures of founder effects, admixture, and selection in the Ashkenazi Jewish population
The paper is free at the PNAS website. The raw data is available. The press release mentions:
In addition, a region on chromosome 12 showed selection in the Ashkenazi Jewish population but not Europeans. This area encompasses 18 genes but the investigators noticed that one of these, ALDH2, is important in alcohol metabolism, and genetic variation in ALDH2 has previously been shown to affect alcohol consumption, Bray says.The abstract refers to ALDH2 as a genomic region under selection that accounts for "alcohol tolerance", but the Jewish version in fact confers alcohol intolerance:
"This is consistent with historical and modern reports of lower alcoholism rates in Jews, although social and religious practices are also thought to play a role," he says. "However, a more detailed analysis of variants in the ALDH2 gene would be necessary to show a mechanistic link."
Reduction in activity of the mitochondrial aldehyde dehydrogenase 2 (ALDH2) enzyme due to genetic deficiency causes reactions related to alcohol consumption and lowers the risk of alcoholism.The authors suggest that the proliferation of the defective ALDH2 version among Jews may be incidental to selection on a different gene:
The mechanism driving selection of the ALDH2 locus is unknown, but a plausible target of selection also within this selected region is the TRAFD1/FLN29 gene, which is a negative regulator of the innate immune system, important for controlling the response to bacterial and viral infection (49). TRAFD1/FLN29 may have conferred a selective advantage in the immune response to a pathogen, perhaps near the time that the Jews returned to Israel from their Babylonian captivity. Despite the unclear selective mechanism, this remains a remarkable example of a putatively selected region accounting for a known population phenotype.Reference: Bray et al. Signatures of founder effects, admixture, and selection in the Ashkenazi Jewish population. PNAS. Published online before print August 26, 2010, doi: 10.1073/pnas.1004381107
Another intra-European AIMs paper
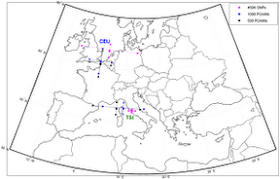
Drineas P, Lewis J, Paschou P (2010) Inferring Geographic Coordinates of Origin for Europeans Using Small Panels of Ancestry Informative Markers. PLoS ONE 5(8): e11892.
Analyzing 1,200 individuals from 11 populations genotyped for more than 500,000 SNPs (Population Reference Sample), we present a systematic exploration of the extent to which geographic coordinates of origin within Europe can be predicted, with small panels of SNPs. Markers are selected to correlate with the top principal components of the dataset, as we have previously demonstrated. Performing thorough cross-validation experiments we show that it is indeed possible to predict individual ancestry within Europe down to a few hundred kilometers from actual individual origin, using information from carefully selected panels of 500 or 1,000 SNPs. Furthermore, we show that these panels can be used to correctly assign the HapMap Phase 3 European populations to their geographic origin. [. . .] It is also worth noting that the largest average error was in the German samples and that the most accurately predicted populations were the Southern European and Irish ones. [. . .] Interestingly, within Europe, individual origin seems much easier to predict along the North to South axis than along the East to West axis. This could indicate increased gene flow along the latter axis.Estimated coordinates for the CEU sample (Utah whites):
Two old papers on Tarim Basin mummies / crania
Now available online at the Sino-Platonic Papers website.
Dolkun Kamberi, "The Three Thousand Year Old Charchan Man" Sino-Platonic Papers, 44 (January, 1994)
HAN Kangxin: "The Study of Ancient Human Skeletons from Xinjiang, China" Sino-Platonic Papers, 5 1 (November, 1994)
Dolkun Kamberi, "The Three Thousand Year Old Charchan Man" Sino-Platonic Papers, 44 (January, 1994)
The male would have been two meters tall when living (see color plates I and IIa); the corpse was lying on its right side with legs bent and propped up by a small piece of wood (perhaps to promote preservation by means of circulation of air around the corpse). The hair, eyelashes, beard and chest hair were intact and traces of makeup (ocher spiral sun-symbols) could be seen on the face. The presence in the tomb of two small bone spoons with dried ocher pigment in them may indicate that the makeup was applied after death. The male's head hair was yellowish brown half gone to white;Note: More recent estimates put Cherchen Man's height closer to 5'9".
HAN Kangxin: "The Study of Ancient Human Skeletons from Xinjiang, China" Sino-Platonic Papers, 5 1 (November, 1994)
Between 1920 and 1940, only three foreign scientists published completed research in this area. They are: Arthur Keith of England (1929), Carl-Herman Hjortsjo and Ander Walander of Germany (1924) and A.N. Iuzefovich of the USSR (1949). A total of twenty skulls were described. Five came from the northern part of the Taklamakan Desert and Keith thought they characterized the "Loulan racial type." Eleven skulls were collected by Sven Hedin from near Luobubo (Lopnor) in 1928 and 1934, and have been subdivided into three groups (Nordic, Chinese, and Alpine) by Hjortsjo and Walander. The remaining skulls also came from the Luobubo (Lopnor) area and exhibit Mongoloid characteristics. Iuzefovich considered these to be of Tujue (Turkish) origin (Keith, A., 1929; Hjortsjo, C.H. and A. Walander, 1942; Iusefovich, A.N., 1949). [. . .]
Chinese scientists have conducted systematic excavations in this region since 1940. I have studied all the skeletal material housed at the Institute of Archeology of Xinjiang and analyzed the physical and racial characteristics of these human bones. The materials included about 274 skulls which were collected from nine ancient cemeteries in Xinjiang. The cemeteries range in age from about 1800 B.C.E. to 300 C.E.
Ancient French megalithic mtDNA
Sample size is three:
We reproducibly retrieved partial HVR-I sequences (nps 16,165 to 16,390) from three human remains (Prisse´ 1, 2, and 4, Table 1), one adult and two children deposited during different stages of use of the burial chamber. Corresponding sequences could be unambiguously assigned to haplogroups X2, U5b, and N1aMarie-France Deguilloux et al. News from the west: Ancient DNA from a French megalithic burial chamber. American Journal of Physical Anthropology DOI: 10.1002/ajpa.21376.
Recent paleogenetic studies have confirmed that the spread of the Neolithic across Europe was neither genetically nor geographically uniform. To extend existing knowledge of the mitochondrial European Neolithic gene pool, we examined six samples of human skeletal material from a French megalithic long mound (c.4200 cal BC). We retrieved HVR-I sequences from three individuals and demonstrated that in the Neolithic period the mtDNA haplogroup N1a, previously only known in central Europe, was as widely distributed as western France. Alternative scenarios are discussed in seeking to explain this result, including Mesolithic ancestry, Neolithic demic diffusion, and long-distance matrimonial exchanges. In light of the limited Neolithic ancient DNA (aDNA) data currently available, we observe that all three scenarios appear equally consistent with paleogenetic and archaeological data. In consequence, we advocate caution in interpreting aDNA in the context of the Neolithic transition in Europe. Nevertheless, our results strengthen conclusions demonstrating genetic discontinuity between modern and ancient Europeans whether through migration, demographic or selection processes, or social practices.
Ezra Pound: racial preservationist
From one of his World War 2 radio speeches:
I am not arguing, I am just telling you. One of these days you will have to start thinking about the problem of race, BREED, preservation.Incidentally, Ezra Pound is another person with more New England ancestry than George Gilder. (Four of Pound's great-grandparents were born in Massachusetts; one was born in New Hampshire; one was born in New York, with ancestry entirely tracing back to Connecticut; one was a Quaker from New Jersey; and one was from Ireland.) Update: More from Pound below.
I do NOT like to think of my race as going toward total extinction, NOR into absolute bondage.
The Cincinnati etc. erred from snobbery. They did not in George Washington’s time organize on racial basis. No one thought of it, no one could have then thought of it. There WAS a racially homogeneous population in the newly freed colonies. Certain privileges were dear to the privileged. Snobbism is NOT conservative. Fashion is not conservative. La Mode, etc. is a ramp.
[#51 (July 2, 1942) U.S.(B65) DISBURSEMENT OF WISDOM]
Intelligence and pigmentation in Hirsch's sample
Additional results from Nathaniel Hirsch's 1926 study of children of immigrants -- this table further subdividing the sample by degree of hair and eye pigmentation.
In this section of the study, Hirsch is opposing some straw man version of Nordicism:
Reference: Hirsch, Nathaniel D. M. A study of natio-racial mental differences. Worcester, Mass. : Clark University, c1926.
In this section of the study, Hirsch is opposing some straw man version of Nordicism:
The Racialists maintain not only that the Nordics are long-headed, tall people with blue eyes and light hair, but more importantly it is claimed that they are mentally superior to all non-Nordic races.And the results satisfy him:
that intellectual differences although (1) significant, (2) innate and (3) relatively permanent are Natio-Racial in character rather than Racial.Which definitely shows "the Racialists". I did find the differences between pigmentation morphotypes within some national groups interesting (though I don't put undue weight on these results), considering light hair and light eyes have apparently been reported elsewhere as correlates of IQ. You can read the entire monograph (which includes more information on the tests used and answers to some criticisms) online at Harvard's open collection on immigration.
Reference: Hirsch, Nathaniel D. M. A study of natio-racial mental differences. Worcester, Mass. : Clark University, c1926.
"Social Darwinism" in Anglophone academic journals
GEOFFREY M. HODGSON. Social Darwinism in Anglophone Academic Journals. Journal of Historical Sociology, 17(4), December 2004, pp. 428-63. (pdf)
This essay is a partial history of the term ‘Social Darwinism’. Using large electronic databases, it is shown that the use of the term in leading Anglophone academic journals was rare up to the 1940s. Citations of the term were generally disapproving of the racist or imperialist ideologies with which it was associated. Neither Herbert Spencer nor William Graham Sumner were described as Social Darwinists in this early literature. Talcott Parsons (1932, 1934, 1937) extended the meaning of the term to describe any extensive use of ideas from biology in the social sciences. Subsequently, Richard Hofstadter (1944) gave the use of the term a huge boost, in the context of a global anti-fascist war.Hodgson mentions:
A massive 1934 fresco by Diego Rivera in Mexico City is entitled ‘Man at the Crossroads’. To the colorful right of the picture are Diego’s chosen symbols of liberation, including Karl Marx, Vladimir Illych Lenin, Leon Trotsky, several young female athletes and the massed proletariat. To the darker left of the mural are sinister battalions of marching gas-masked soldiers, the ancient statue of a fearsome god, and the seated figure of a bearded Charles Darwin. These conceptions of good and evil, progress and regress, and light and shade, were prominent in much of Western social science for the next fifty years.Diego Rivera attributed his politics to his Jewish ancestry:
"My Jewishness is the dominant element in my life," Rivera wrote in 1935. "From this has come my sympathy with the downtrodden masses which motivates all my work."Coincidentally (I'm sure), Hofstadter was also a Jewish-identified half-Jew. More excerpts within: